Tonga Volcano Eruption Caused Massive Space Plasma Disturbances on a Global Scale
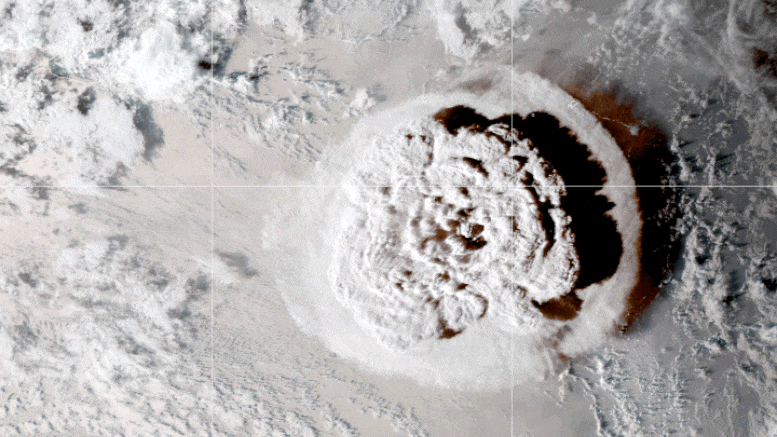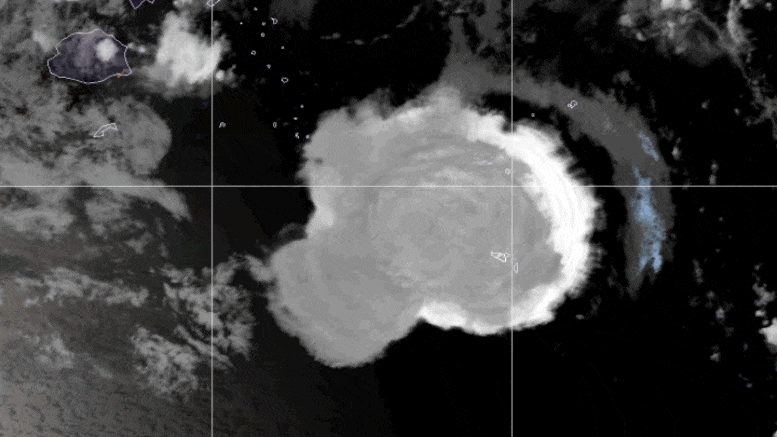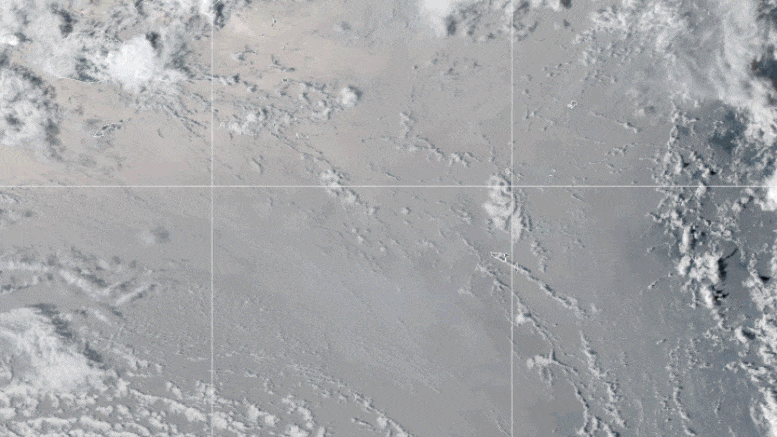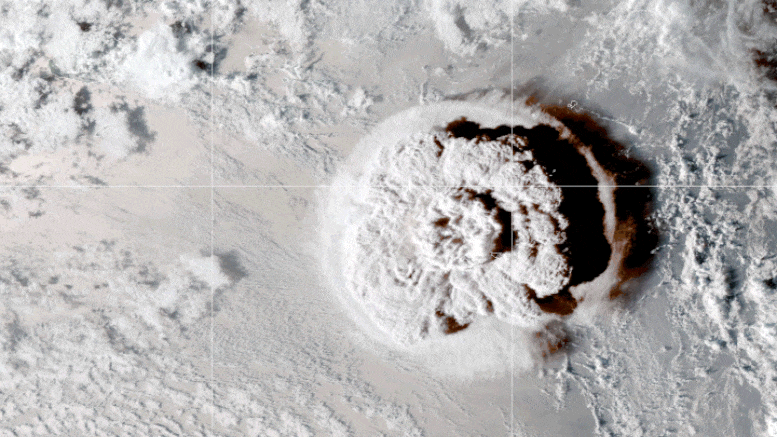
This looping video shows a series of GOES-17 satellite images that caught an umbrella cloud generated by the underwater eruption of the Hunga Tonga-Hunga Ha’apai volcano on January 15, 2022. Crescent-shaped bow shock waves and numerous lighting strikes are also visible. Credit: NASA Earth Observatory image by Joshua Stevens using GOES imagery courtesy of NOAA and NESDIS
MIT Haystack Observatory identifies long-duration atmospheric waves launched by the recent Tonga eruption.
The recent eruption of Tonga’s Hunga Tonga–Hunga Ha‘apai volcano, at 04:14:45 UT on January 15, 2022, was recently confirmed to have launched far-reaching, massive global disturbances in the Earth’s atmosphere.
Using data recorded by more than 5,000 Global Navigation Satellite System (GNSS) ground receivers located around the globe, MIT Haystack Observatory scientists and their international partners from the Arctic University of Norway have observed substantial evidence of eruption-generated atmospheric waves and their ionospheric imprints 300 kilometers above the Earth’s surface over an extended period. These atmospheric waves were active for at least four days after the eruption and circled the globe three times. Ionospheric disturbances passed over the United States six times, at first from west to east and later in reverse.
This volcanic event was extraordinarily powerful, releasing energy equivalent to 1,000 atomic bombs of the size deployed in 1945. Scientists have known that explosive volcanic eruptions and earthquakes can trigger a series of atmospheric pressure waves, including acoustic waves, and that they can perturb the upper atmosphere a few hundred kilometers above the epicenter. When over the ocean, they can trigger tsunami waves, and therefore upper-atmospheric disturbances. Results from this Tonga eruption have surprised this international team, particularly in their geographic extent and multiple-day durations. These discoveries ultimately suggest new ways in which the atmospheric waves and the global ionosphere are connected.
A new study, led by researchers at MIT Haystack Observatory and the Arctic University of Norway, reporting the results was published on March 23, 2022, in the peer-reviewed journal Frontiers in Astronomy and Space Sciences.
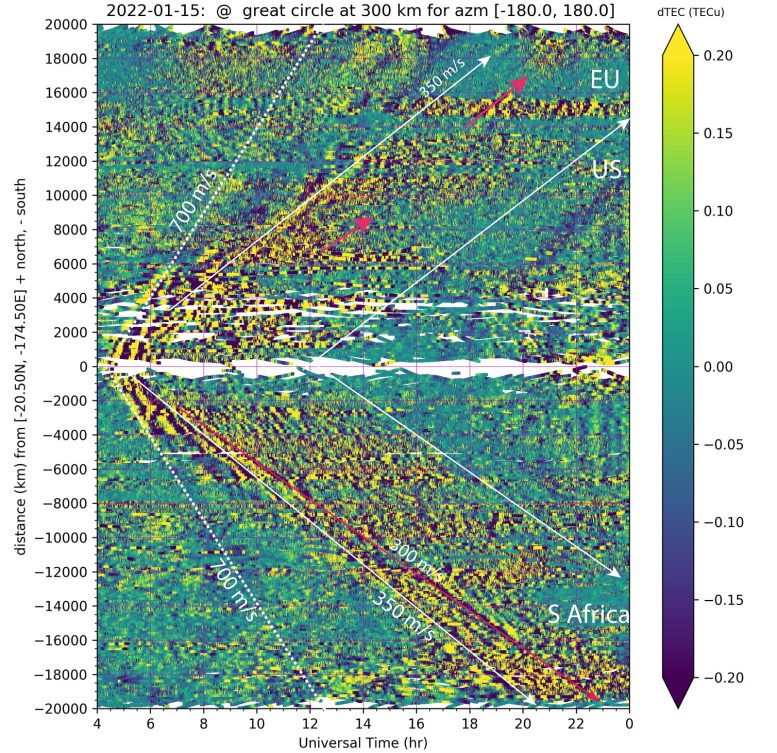
Traveling ionospheric disturbances following an eruption in the kingdom of Tonga in the South Pacific Ocean, as measured from the global GNSS networks of receivers. The horizontal axis shows time; the vertical axis shows distance. Yellow areas within the white-line envelope as marked by fiducial arrows are enhanced ionospheric disturbances in total electron content (TEC). Distance is measured along Great-Circle loci with origin at Tonga. The positive and negative distance shows TIDs propagating both northward and southward from Tonga. The eruption antipode is in North Africa, approximately 21,000 km away from Tonga. TIDs took 17-18 hours to reach the antipode and the same time to return to Tonga on the next day. Credit: Shunrong Zhang/Haystack Observatory
The authors believe the disturbances to be an effect of Lamb waves; these waves, named after mathematician Horace Lamb, travel at the speed of sound globally without much reduction in amplitude. Although they are located predominantly near Earth’s surface, these waves can exchange energy with the ionosphere through complex pathways. As stated in the new paper, “prevailing Lamb waves have been reported before as atmospheric responses to the Krakatoa eruption in 1883 and other geohazards. This study provides substantial first evidence of their long-duration imprints up in the global ionosphere.”
Under National Science Foundation support, Haystack has been assembling global GNSS network observations to study important total electron content information on a daily basis since 2000. The observatory shares this data with the international geospace community to enable innovative research on a variety of frontiers, ranging from solar storm effects to low atmospheric forcing. A particular form of space weather, caused by ionospheric waves called traveling ionospheric disturbances (TIDs), are often excited by processes including sudden energy inputs from the sun, terrestrial weather, and human-made disturbances. For example, Haystack scientists used TID observations to provide the first evidence that solar eclipses can trigger bow waves in Earth’s atmosphere.
Lead author Shunrong Zhang says, “Only severe solar storms are known to produce TID global propagation in space for several hours, if not for days; volcanic eruptions and earthquakes normally yield ionospheric disturbances only within thousands of kilometers. By detecting these significant eruption-induced ionospheric disturbances in space over very large distances, we found not only generation of Lamb waves and their global propagation over several days (often monitored as sound waves on the ground for compliance with Comprehensive Nuclear Test Ban Treaties) but also a fundamental new physical process. In the end, surface and lower atmospheric signals can make a loud splash, even deep in space.”
Beyond these results, Haystack scientists continue additional studies of the Tonga eruption’s generation of severe space weather effects.
Reference: “2022 Tonga Volcanic Eruption Induced Global Propagation of Ionospheric Disturbances via Lamb Waves” by Shun-Rong Zhang, Juha Vierinen2, Ercha Aa, Larisa P. Goncharenko, Philip J. Erickson, William Rideout, Anthea J. Coster and Andres Spicher, 23 March 2022, Frontiers in Astronomy and Space Sciences.
DOI: 10.3389/fspas.2022.871275
Describing the devastating eruption in Tonga
Just two months after the eruption, geologists have put together a preliminary account of how it unfolded. UC Santa Barbara's Melissa Scruggs and emeritus Professor Frank Spera were part of an international team of researchers that published the first holistic account of the event in the journal Earthquake Research Advances. The authors think that an eruption the day before may have primed the volcano for the violent explosion by sinking its main vent below the ocean's surface. This enabled molten rock to vaporize a large volume of seawater, intensifying the volcanic eruption the very next day.
"This is definitely, without a doubt, the largest eruption since Mt. Pinatubo in 1991," said corresponding author Scruggs, who studies magma mixing and eruption triggering mechanisms, and recently completed her doctorate at UC Santa Barbara. She compared January's event to the 1883 eruption of Krakatoa, which was heard 3,000 miles away.
Hunga Tonga-Hunga Ha'apai (HTHH) is a stratovolcano: a large, cone-shaped mountain that is prone to periodic violent eruptions, but which usually experiences milder activity. It's one of many along the Tofua Volcanic Arc, a line of volcanoes fed by magma from the Pacific Plate diving beneath the Indo-Australian Plate. Heat and pressure cook the rocks of the descending plate, driving out water and other volatiles. That same water decreases the melting temperature of the rock above, leading to a chain of volcanoes about 100 kilometers from the plate boundary.
A submerged danger
The islands of Hunga Tonga and Hunga Ha'apai -- after which the volcano is named -- are merely the two highest points along the rim of the caldera, or central crater. Or they were, until the eruption blew most of the islands sky high.
Scruggs first heard about the eruption as she scrolled through her Twitter feed while getting ready for bed. "I saw a GIF of the satellite eruption, and my heart just stopped," she said, pausing to find her words. She immediately knew that the event would cause massive devastation. "The scariest part was that the entire country was cut off, and we didn't know what had happened."
She was already messaging other volcanologists as the events unfolded, trying to understand the images that satellites had so clearly captured. "We really just set out to try to understand what happened," Scruggs said. "So, we gathered all the information that we could, anything that was available within the first few weeks." The authors drew on whatever resources they could find to quickly characterize this eruption, including publicly available data, videos and even tweets.
Using a variety of data sets, the team calculated that the January 15 event began at 5:02 p.m. local time (0402 ±1 UTC). The U.S. Geological Survey recorded a seismic event around 13 minutes later at the vent location. The first two hours of the eruption were particularly violent, with activity fading after about 12 hours.
But eruption activity had actually started all the way back on December 20, 2021. And before that, the volcano had erupted in 2009 and again in 2014 and 2015. Scruggs believes these earlier episodes are key to understanding the violence behind HTHH's recent eruption, perhaps related to changes in the magma plumbing system at depth or the chemistry of the magma over time.
Hunga Tonga and Hunga Ha'apai used to be separate islands until they were united by eruptions from the volcano's main vent, which created a land bridge. "This island was just born in 2015," said Scruggs. "And now it's gone. Were it not for the satellite era, we would not have even known it ever existed."
On January 14, 2022 an explosion from the main vent razed this connection, sinking the vent beneath the ocean's surface. "Had that land bridge not been taken out, the January 15 eruption might have behaved just like the day before because it would not have had that excess seawater," Scruggs remarked.
A staggering explosion
Same volcano, one day's difference: On Friday the vent was above the water, and by Saturday it was below. "That made all of the difference in the world," Scruggs said.
The team believes that the seawater played a large part in the violence and force behind the Jan. 15 eruption. Much like a bottle rocket, an eruption of this scale takes the right ratio of water and gas to provide the force to send it skyward.
And it took off like a rocket, too. "It went halfway to space," Scruggs exclaimed. The ash plume shot 58 kilometers into the atmosphere, past the stratosphere and into the lower mesosphere. This is more than twice the height reached by the plume from Mt. Saint Helens in 1980. It was the tallest volcanic plume ever recorded.
A truly staggering amount of lightning also accompanied the eruption. The authors suspect that vaporizing seawater caused the lava to fragment into microscopic ash particles, which were joined by tiny ice crystals once the steam froze in the upper atmosphere. The motion, temperature change and size of the particles generated incredible amounts of static charge separation that flashed above the eruption. For the first two hours of the eruption, about 80% of all lightning strikes on Earth split the sky above Hunga Tonga-Hunga Ha'apai.
The authors estimate around 1.9 km3 of material, weighing 2,900 teragrams, erupted from HTHH on Jan. 15. "But the volume of the eruption was not the big deal," said Spera, a coauthor on the paper and Scruggs' doctoral advisor. "What was special is how the energy of the eruption coupled to the atmosphere and oceans: A lot of the energy went into moving air and water on a global scale."
The shockwave traveling through the ocean triggered tsunamis throughout the Pacific, and beyond. What's more, the wave arrived faster than tsunami warning models predicted because the models aren't calibrated for volcanic eruptions -- they're based on equations that describe tsunamis generated by earthquakes.
A second tsunami followed the atmospheric pressure wave. This shockwave even triggered a meteo-tsunami in the Caribbean, which has no direct connection to the South Pacific. Scruggs called it unprecedented: "Basically the whole ocean just kind of sloshed around for five days after the eruption," she added.
Plenty of work to do
Scientists are still piecing together what happened at the volcano, so they have yet to develop a complete understanding of the tsunami wave. However, it's an important task needed to update tsunami travel forecast systems so they account for this type of mechanism. Otherwise, warnings could be incorrect the next time a volcano like HTHH erupts, potentially costing more lives.
Indeed, the event highlights the danger posed by unmonitored submarine volcanoes. Despite the devastation, the people of Tonga were relatively well prepared for the Jan. 15 eruption. The government had issued warnings based on the previous day's activity, and the nation had plans in place for eruptions and tsunamis.
HTHH has experienced similarly violent eruptions in the past. A recent paper by researchers at the University of Otago, New Zealand revealed that a large eruption destroyed the caldera at the summit of the undersea volcano about 1,000 years ago. And similar volcanoes could well erupt in the same manner. Consider Kick 'em Jenny, another submarine volcano whose main vent is a mere 150 meters underwater. It's located just 8 km north of the island of Grenada. "Imagine if something like the Tonga eruption happened in the Caribbean," Scruggs said.
The researchers worked quickly with only publicly available data. They plan to revisit all their findings as more information and samples become available and as more researchers publish their own findings on this groundbreaking eruption. Their primary goal was to provide a point of departure for future work on the topic.
Scruggs is particularly keen on learning about the ash collected from this eruption. Volcanic rock provides a wealth of information to a trained geologist. Examining the material could shed light on the type of magma that erupted, how much of it there was and perhaps even how much seawater was involved in the eruption.
"There's so many questions that have been raised," said Scruggs. "Things we didn't even think were possible have now been recorded."
The UC Santa Barbara researchers will lead a special invited session on the Hunga Tonga-Hunga Ha'apai eruption at the Geological Society of America's 2022 annual meeting in Denver this October. "It will be exciting to see what scores of other earth scientists can discover about this unique volcano," Spera said. "We are just at the beginning."
Journal Reference:
David A. Yuen, Melissa A. Scruggs, Frank J. Spera, Yingcai Zheng, Hao Hu, Stephen R. McNutt, Glenn Thompson, Kyle Mandli, Barry R. Keller, Songqiao Shawn Wei, Zhigang Peng, Zili Zhou, Francesco Mulargia, Yuichiro Tanioka. Under the Surface: Pressure-Induced Planetary-Scale Waves, Volcanic Lightning, and Gaseous Clouds Caused by the Submarine Eruption of Hunga Tonga-Hunga Ha’apai Volcano Provide an Excellent Research Opportunity. Earthquake Research Advances, 2022; 100134 DOI: 10.1016/j.eqrea.2022.100134