An open access software-based tool for predicting COVID-19 susceptibility in animals
The world is currently in the grip of the COVID-19 pandemic. Preventing the spread of COVID-19 from human to wildlife and domestic animals is an immediate concern of medical scientists worldwide. Recently, there have been reports of cats, ferrets, dogs, minks, golden hamsters, rhesus monkeys, tigers and lions testing positive for SARS-CoV-2 RNA. Preventing the spread of COVID-19 in the zoo and wildlife sanctuaries where chances of animal-human contact are higher should be a priority, as disease may threaten many rare and near-extinct animal species. Additionally, the continued presence of the virus in animals can work as a reservoir that can be a potential reason for failure in containment and recurrence of the pandemic.
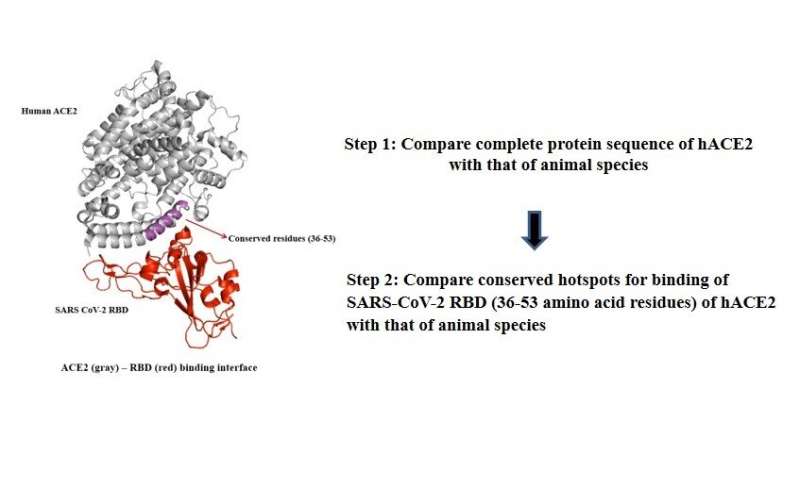
SARS-CoV-2 infects humans using a cell-surface protein—angiotensin converting enzyme-2 (ACE2). The receptor binding domain (RBD) of SARS-CoV-2 binds to N-terminal end of ACE2, which is a key step for host cell entry of the virus. The animals that have the ACE2 protein sequence similar to humans, specifically for the segment of protein which binds to SARS-CoV-2 RBD, may also become infected with this virus.
Using the open-access NCBI protein database and protein blast tools, our research group, the Etiologically Elusive Disorders Research Network (EEDRN), has devised a two-step bioinformatics-based method for comparing the homology of human ACE2 with that of animal species-specific ACE2 protein sequence. First, we performed a comparative analysis of the variability of hACE2 with that of wildlife and domestic animal species in complete protein sequences. The species that showed significant homology for the complete sequence were selected for further analysis for the second step.
In the second step, we narrowed down our homology search to a sub-range of amino acid residues of human ACE2, which contain conserved hotspots for binding of SARS‐CoV‐2 RBD. The susceptibility for contracting SARS-CoV-2, and the degree of risk of infection for any animal species for which the ACE2 protein sequence is available at NCBI database can be easily calculated online in these two simple steps using open access NCBI protein blast tools.
On the basis of the sequence homology, we predicted the highest susceptibility for hominids (such as chimpanzee, gorilla, and rhesus monkey) and other primates, followed by carnivores, rodents and artiodactyles (ungulates). The risk for contracting the virus showed a distinct evolutionary trend—the closer to humans in evolution, the higher is the risk of infection. We tested our predictions against PCR‐based laboratory testing results for SARS‐CoV‐2 RNA reported in recent literature for animals such as cats, dogs, golden hamster, rhesus monkey, tigers and lions, which showed our results were highly accurate. Our proposed method can be used as a no-cost screening tool for guiding viral RNA testing for domestic and wildlife animals at risk of getting COVID-19, especially at the settings where immediate availability of PCR-based testing facility cannot be assured.
This story is part of Science X Dialog, where researchers can report findings from their published research articles. Visit this page for information about ScienceX Dialog and how to participate.
More information: Kumar A, Pandey SN, Pareek V, Narayan RK, Faiq MA, Kumari C. Predicting susceptibility for SARS‐CoV‐2 infection in domestic and wildlife animals using ACE2 protein sequence homology. Zoo Biology. 2020;1–7. doi.org/10.1002/zoo.21576.
Bio:
Dr. Ashutosh Kumar is an assistant professor and Dr. Ravi K. Narayan is a senior resident doctor, at Department of Anatomy, All India Institute of Medical Sciences (AIIMS), Patna, India.
No comments:
Post a Comment