A common gene-editing enzyme could be used to disable RNA viruses such as flu or Ebola
By Tanya Lewis on October 23, 2019
Researchers modified the enzyme Cas13 to target and inactivate viruses such as influenza (shown here). Credit: Kateryna Kon Getty Images
CRISPR is usually thought of as a laboratory tool to edit DNA in order to fix genetic defects or enhance certain traits—but the mechanism originally evolved in bacteria as a way to fend off viruses called bacteriophages. Now scientists have found a way to adapt this ability to fight viruses in human cells.
In a recent study, Catherine Freije, Cameron Myhrvold and Pardis Sabeti at the Broad Institute of the Massachusetts Institute of Technology and Harvard University, and their colleagues programmed a CRISPR-related enzyme to target three different single-stranded RNA viruses in human embryonic kidney cells (as well as human lung cancer cells and dog kidney cells) grown in vitro and chop them up, rendering them largely unable to infect additional cells. If further experiments show this process works in living animals, it could eventually lead to new antiviral therapies for diseases such as Ebola or Zika in humans.
Viruses come in many forms, including DNA and RNA, double-stranded and single-stranded. About two thirds of the ones that infect humans are RNA viruses, and many have no approved treatment. Existing therapies often use a small molecule that interferes with viral replication—but this approach does not work for newly emerging viruses or ones that are evolving rapidly.
“CRISPR” refers to a series of DNA sequences in bacterial genomes that were left behind from previous bacteriophage infections. When the bacteria encounter these pathogens again, enzymes called CRISPR-associated (Cas) proteins recognize and bind to these sequences in the virus and destroy them. In recent years, researchers (including study co-author Feng Zhang) have reengineered one such enzyme, called Cas9, to cut and paste DNA in human cells. The enzyme binds to a short genetic tag called a guide RNA, which directs the enzyme to a particular part of the genome to make cuts. Previous studies have used Cas9 to prevent replication of double-stranded DNA viruses or of single-stranded RNA viruses that produce DNA in an intermediate step during replication. Only a small fraction of RNA viruses that infect humans produce such DNA intermediates—but another CRISPR enzyme, called Cas13, can be programmed to cleave single-stranded RNA viruses.
“The nice thing about CRISPR systems and systems like Cas13 is that their initial purpose in bacteria was to defend against viral infection of bacteria, and so we sort of wanted to bring Cas13 back to its original function—and apply this to mammalian viruses in mammalian cells,” says Freije, who is a doctoral student in virology at Harvard. “Because CRISPR systems rely on guide RNAs to specifically guide the CRISPR protein to a target, we saw this as a great opportunity to use it as a programmable antiviral.”
Freije and her colleagues programmed Cas13 to target three different viruses: lymphocytic choriomeningitis virus (LCMV), influenza A virus (IAV) and vesicular stomatitis virus (VSV). LCMV is an RNA virus that mostly infects mice—but it is in the same family as the virus that causes Lassa fever, which is found in West Africa and is much more dangerous to study in the lab. IAV is a flu virus; although some antiviral medications for flu already exist, such viruses evolve rapidly, so there is a need for better options. Finally, VSV is a model for many other single-stranded RNA viruses.
To determine how effective Cas13 was at destroying the viruses, the researchers also used it as a diagnostic tool to see how much viral RNA was being released from infected cells. They saw a twofold to 44-fold reduction in RNA, depending on which virus they were looking at and the time point. They also looked at how well the released RNA was able to go on and infect new cells. In most cases, they saw a 100-fold reduction in infectivity—and in some cases, more than 300-fold—according to Freije. The findings were published online on October 10 in Molecular Cell.
“The results are very impressive,” says Chen Liang, a professor at the Lady Davis Institute at Jewish General Hospital and the department of microbiology and immunology at McGill University in Montreal, who was not involved in the study. His own laboratory has used the Cas9 enzyme to deactivate DNA viruses. The concept is very similar, but Cas13 has a few advantages, he says. For one, Cas13 can be used to target one virus using several guide RNAs, making it difficult for the virus to “escape.” Secondly, the new study also used Cas13 to detect how much viral RNA was left over to infect cells. The amount of viral knockdown the group achieved is “very significant,” Liang says. “If you can target and inactivate all three [of these] viruses, in principle, you can inactivate any virus.
As with any approach, there are limitations. One is the question of how to deliver the Cas13 to target a virus in a living person, Liang notes, and the researchers have not yet done any animal studies. Another is the fact that viruses will eventually develop resistance. But Cas13 has an advantage here: when Cas9 cuts viral DNA, mammalian cells repair it and can cause mutations that make the virus more resistant. Yet with Cas13, these cells do not have the mechanism to repair the RNA and introduce errors that would help the virus escape being destroyed. Even if a virus does evolve resistance, or if a new virus is encountered, the method could be quickly adapted.
“One of the things that’s most exciting about this approach is the programmability,” says Myhrvold, a postdoctoral fellow at Harvard. “Once you figure out how to do this well for one virus it’s not that hard to design sequences against another virus—or another one. Furthermore, if the virus changes its own sequence—as viruses are known to do, just during an outbreak or in response to therapy—you can very easily update the CRISPR RNA sequence and keep up with the virus.”
Freije agrees. “We are definitely excited about future prospects of optimizing the system and trying it out in mouse models,” she says. Beyond therapeutics, the team hopes to understand more about how viruses operate—how they replicate and what parts of their genomes are most important. Using approaches like this, “you can really start to get a better picture of what parts of these viruses are and, most importantly, what really makes them tick.”
“One of the things that’s most exciting about this approach is the programmability,” says Myhrvold, a postdoctoral fellow at Harvard. “Once you figure out how to do this well for one virus it’s not that hard to design sequences against another virus—or another one. Furthermore, if the virus changes its own sequence—as viruses are known to do, just during an outbreak or in response to therapy—you can very easily update the CRISPR RNA sequence and keep up with the virus.”
Freije agrees. “We are definitely excited about future prospects of optimizing the system and trying it out in mouse models,” she says. Beyond therapeutics, the team hopes to understand more about how viruses operate—how they replicate and what parts of their genomes are most important. Using approaches like this, “you can really start to get a better picture of what parts of these viruses are and, most importantly, what really makes them tick.”
How do bacteria defend themselves against viruses?
The CRISPR-Cas system in some bacteria helps to form an effective barrier to invading viruses.
DIGEST Apr 3, 2019
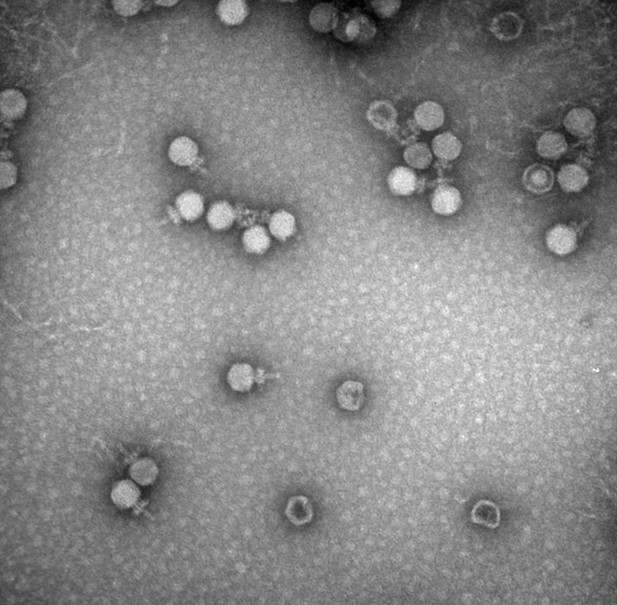
A transmission electron microscopy image of bacteriophages taken at The University of Alabama’s Optical Analysis Facility. Image credit: Chou-Zheng and Hatoum-Aslan, 2019 (CC BY 4.0)
Just as humans are susceptible to viruses, bacteria have their own viruses to contend with. These viruses – known as phages – attach to the surface of bacterial cells, inject their genetic material, and use the cells’ enzymes to multiply while destroying their hosts.
To defend against a phage attack, bacteria have evolved a variety of immune systems. For example, when a bacterium with an immune system known as CRISPR-Cas encounters a phage, the system creates a ‘memory’ of the invader by capturing a small snippet of the phage’s genetic material. The pieces of phage DNA are copied into small molecules known as CRISPR RNAs, which then combine with one or more Cas proteins to form a group called a Cas complex. This complex patrols the inside of the cell, carrying the CRISPR RNA for comparison, similar to the way a detective uses a fingerprint to identify a criminal. Once a match is found, the Cas proteins chop up the invading genetic material and destroy the phage.
There are several different types of CRISPR-Cas systems. Type III systems are among the most widespread in nature and are unique in that they provide a nearly impenetrable barrier to phages attempting to infect bacterial cells. Medical researchers are exploring the use of phages as alternatives to conventional antibiotics and so it is important to find ways to overcome these immune responses in bacteria. However, it remains unclear precisely how Type III CRISPR-Cas systems are able to mount such an effective defense.
Chou-Zheng and Hatoum-Aslan used genetic and biochemical approaches to study the Type III CRISPR-Cas system in a bacterium called Staphylococcus epidermidis. The experiments showed that two enzymes called PNPase and RNase J2 played crucial roles in the defense response triggered by the system. PNPase helped to generate CRISPR RNAs and both enzymes were required to help to destroy genetic material from invading phages.
Previous studies have shown that PNPase and RNase J2 are part of a machine in bacterial cells that usually degrades damaged genetic material. Therefore, these findings show that the Type III CRISPR-Cas system in S. epidermidis has evolved to coordinate with another pathway to help the bacteria survive attack from phages. CRISPR-Cas immune systems have formed the basis for a variety of technologies that continue to revolutionize genetics and biomedical research. Therefore, along with aiding the search for alternatives to antibiotics, this work may potentially inspire the development of new genetic technologies in the future.
Phage-Encoded Anti-CRISPR Defenses
Annual Review of Genetics
Vol. 52:445-464 (Volume publication date November 2018)
First published as a Review in Advance on September 12, 2018
https://doi.org/10.1146/annurev-genet-120417-031321
Sabrina Y. Stanley1 and Karen L. Maxwell2
1Department of Molecular Genetics, University of Toronto, Toronto, Ontario M5S 1A8, Canada
2Department of Biochemistry, University of Toronto, Toronto, Ontario M5G 1M1, Canada; email: karen.maxwell@utoronto.ca
Abstract
The battle for survival between bacteria and bacteriophages (phages) is an arms race where bacteria develop defenses to protect themselves from phages and phages evolve counterstrategies to bypass these defenses. CRISPR-Cas adaptive immune systems represent a widespread mechanism by which bacteria protect themselves from phage infection. In response to CRISPR-Cas, phages have evolved protein inhibitors known as anti-CRISPRs. Here, we describe the discovery and mechanisms of action of anti-CRISPR proteins. We discuss the potential impact of anti-CRISPRs on bacterial evolution, speculate on their evolutionary origins, and contemplate the possible next steps in the CRISPR-Cas evolutionary arms race. We also touch on the impact of anti-CRISPRs on the development of CRISPR-Cas-based biotechnological tools.
The battle for survival between bacteria and bacteriophages (phages) is an arms race where bacteria develop defenses to protect themselves from phages and phages evolve counterstrategies to bypass these defenses. CRISPR-Cas adaptive immune systems represent a widespread mechanism by which bacteria protect themselves from phage infection. In response to CRISPR-Cas, phages have evolved protein inhibitors known as anti-CRISPRs. Here, we describe the discovery and mechanisms of action of anti-CRISPR proteins. We discuss the potential impact of anti-CRISPRs on bacterial evolution, speculate on their evolutionary origins, and contemplate the possible next steps in the CRISPR-Cas evolutionary arms race. We also touch on the impact of anti-CRISPRs on the development of CRISPR-Cas-based biotechnological tools.
FULL ARTICLE HERE
Biological Sciences
RESEARCH ARTICLE
Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria
Ido Yosef, Miriam Manor, Ruth Kiro, and View ORCID Profile Udi Qimron
PNAS June 9, 2015 112 (23) 7267-7272; first published May 18, 2015 https://doi.org/10.1073/pnas.1500107112
Edited by Jennifer A. Doudna, University of California, Berkeley, CA, and approved April 28, 2015 (received for review January 25, 2015)
Significance
Antibiotic resistance of pathogens is a growing concern to human health, reviving interest in phage therapy. This therapy uses phages (natural bacterial enemies) to kill pathogens. However, it encounters many obstacles such as delivery barriers into the tissues and bacterial resistance to phages. Here, we use phages for delivering a programmable DNA nuclease, clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas), to reverse antibiotic resistance and eliminate the transfer of resistance between strains. This approach combines CRISPR-Cas delivery with lytic phage selection of antibiotic-sensitized bacteria. The strategy may reduce the prevalence of antibiotic-resistant bacteria in treated surfaces and on skin of medical personnel, as it uses phages in a unique way that overcomes many of the hurdles encountered by phage therapy.
Abstract
The increasing threat of pathogen resistance to antibiotics requires the development of novel antimicrobial strategies. Here we present a proof of concept for a genetic strategy that aims to sensitize bacteria to antibiotics and selectively kill antibiotic-resistant bacteria. We use temperate phages to deliver a functional clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas) system into the genome of antibiotic-resistant bacteria. The delivered CRISPR-Cas system destroys both antibiotic resistance-conferring plasmids and genetically modified lytic phages. This linkage between antibiotic sensitization and protection from lytic phages is a key feature of the strategy. It allows programming of lytic phages to kill only antibiotic-resistant bacteria while protecting antibiotic-sensitized bacteria. Phages designed according to this strategy may be used on hospital surfaces and hand sanitizers to facilitate replacement of antibiotic-resistant pathogens with sensitive ones.
Biological Sciences
RESEARCH ARTICLE
Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria
Ido Yosef, Miriam Manor, Ruth Kiro, and View ORCID Profile Udi Qimron
PNAS June 9, 2015 112 (23) 7267-7272; first published May 18, 2015 https://doi.org/10.1073/pnas.1500107112
Edited by Jennifer A. Doudna, University of California, Berkeley, CA, and approved April 28, 2015 (received for review January 25, 2015)
Significance
Antibiotic resistance of pathogens is a growing concern to human health, reviving interest in phage therapy. This therapy uses phages (natural bacterial enemies) to kill pathogens. However, it encounters many obstacles such as delivery barriers into the tissues and bacterial resistance to phages. Here, we use phages for delivering a programmable DNA nuclease, clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas), to reverse antibiotic resistance and eliminate the transfer of resistance between strains. This approach combines CRISPR-Cas delivery with lytic phage selection of antibiotic-sensitized bacteria. The strategy may reduce the prevalence of antibiotic-resistant bacteria in treated surfaces and on skin of medical personnel, as it uses phages in a unique way that overcomes many of the hurdles encountered by phage therapy.
Abstract
The increasing threat of pathogen resistance to antibiotics requires the development of novel antimicrobial strategies. Here we present a proof of concept for a genetic strategy that aims to sensitize bacteria to antibiotics and selectively kill antibiotic-resistant bacteria. We use temperate phages to deliver a functional clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated (Cas) system into the genome of antibiotic-resistant bacteria. The delivered CRISPR-Cas system destroys both antibiotic resistance-conferring plasmids and genetically modified lytic phages. This linkage between antibiotic sensitization and protection from lytic phages is a key feature of the strategy. It allows programming of lytic phages to kill only antibiotic-resistant bacteria while protecting antibiotic-sensitized bacteria. Phages designed according to this strategy may be used on hospital surfaces and hand sanitizers to facilitate replacement of antibiotic-resistant pathogens with sensitive ones.
DOWNLOAD WHOLE PAPER AS PDF FROM HERE
SUMMARY

Volume 366
Issue 9
May 2019
Article Contents
ABSTRACT
INTRODUCTION
BIOLOGICAL RELEVANCE OF ANTI-CRISPR PROTEINS
MECHANISMS AND STRUCTURES OF ANTI-CRISPR PROTEINS
APPLICATIONS OF ANTI-CRISPR PROTEINS
OUTLOOK
FUNDING
REFERENCES
MINI REVIEW
Keeping CRISPR in check: diverse mechanisms of phage-encoded anti-CRISPRS
Despoina Trasanidou, Ana Sousa Gerós, Prarthana Mohanraju, Anna Cornelia Nieuwenweg, Franklin L Nobrega, Raymond H J Staals
FEMS Microbiology Letters, Volume 366, Issue 9, May 2019, fnz098, https://doi.org/10.1093/femsle/fnz098
Published: 11 May 2019
ABSTRACT
CRISPR-Cas represents the only adaptive immune system of prokaryotes known to date. These immune systems are widespread among bacteria and archaea, and provide protection against invasion of mobile genetic elements, such as bacteriophages and plasmids. As a result of the arms-race between phages and their prokaryotic hosts, phages have evolved inhibitors known as anti-CRISPR (Acr) proteins to evade CRISPR immunity. In the recent years, several Acr proteins have been described in both temperate and virulent phages targeting diverse CRISPR-Cas systems. Here, we describe the strategies of Acr discovery and the multiple molecular mechanisms by which these proteins operate to inhibit CRISPR immunity. We discuss the biological relevance of Acr proteins and speculate on the implications of their activity for the development of improved CRISPR-based research and biotechnological tools.

The physicist's guide to one of biotechnology's hottest new topics: CRISPR-Cas
Melia E Bonomo1,3 and Michael W Deem1,2,3,4
Published 30 April 2018 • © 2018 IOP Publishing Ltd
Physical Biology, Volume 15, Number 4
DownloadArticle PDF
Article information
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) constitute a multi-functional, constantly evolving immune system in bacteria and archaea cells. A heritable, molecular memory is generated of phage, plasmids, or other mobile genetic elements that attempt to attack the cell. This memory is used to recognize and interfere with subsequent invasions from the same genetic elements. This versatile prokaryotic tool has also been used to advance applications in biotechnology. Here we review a large body of CRISPR-Cas research to explore themes of evolution and selection, population dynamics, horizontal gene transfer, specific and cross-reactive interactions, cost and regulation, non-immunological CRISPR functions that boost host cell robustness, as well as applicable mechanisms for efficient and specific genetic engineering. We offer future directions that can be addressed by the physics community. Physical understanding of the CRISPR-Cas system will advance uses in biotechnology, such as developing cell lines and animal models, cell labeling and information storage, combatting antibiotic resistance, and human therapeutics.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
In 1987, Ishino and colleagues had set out to identify the encoded protein and primary structure of a particular gene in Escherichia coli by analyzing its chromosomal DNA segment and flanking regions [1]. They found an interesting sequence structure at the gene's 3'-end flanking region, in which five homologous sequences of 29 nucleotides were arranged as direct repeats with 32-nucleotide sequences spaced between them. Little did they know that their discovery would prove to have critical immunological significance. It was not until 2000 that these mysterious repeated genomic elements were revisited when Mojica and colleagues searched the available microbial genome database and found many organisms that contained partially palindromic sequences of 24–40 basepairs with 20–58 basepair sequences spaced between them [2]. These were found in almost all archaea, about half of bacteria, no viruses, and no eukaryotes. Related and unrelated species had nearly identical structure in these repeat sequence units. The sequences in between, called 'spacers', were unique to an individual locus and were not found in other genomes [3]. After many suggested abbreviations, including SRSRs, short regularly spaced repeats, and SPIDR, spacers interspersed direct repeats, the scientific community settled on calling these elements clustered regularly interspaced short palindromic repeats, or CRISPR.
SEE
https://plawiuk.blogspot.com/search?q=BACTERIOPHAGES
https://plawiuk.blogspot.com/search?q=PHAGES
Genetically Engineered Phages: a Review of Advances over the Last Decade
DOI: 10.1128/MMBR.00069-15
SUMMARY
Soon after their discovery in the early 20th century, bacteriophages were recognized to have great potential as antimicrobial agents, a potential that has yet to be fully realized. The nascent field of phage therapy was adversely affected by inadequately controlled trials and the discovery of antibiotics. Although the study of phages as anti-infective agents slowed, phages played an important role in the development of molecular biology. In recent years, the increase in multidrug-resistant bacteria has renewed interest in the use of phages as antimicrobial agents. With the wide array of possibilities offered by genetic engineering, these bacterial viruses are being modified to precisely control and detect bacteria and to serve as new sources of antibacterials. In applications that go beyond their antimicrobial activity, phages are also being developed as vehicles for drug delivery and vaccines, as well as for the assembly of new materials. This review highlights advances in techniques used to engineer phages for all of these purposes and discusses existing challenges and opportunities for future work.
INTRODUCTION
Bacteriophages (phages) are among the most abundant biological particles on earth. They are also highly versatile and adaptable to a great number of applications. Phages are viruses that infect bacteria; their self-replication depends on access to a bacterial host. Phages were discovered independently by Frederick Twort in 1915 (1) and by Félix d'Hérelle in 1917 (2), and they were used early on as antimicrobial agents. Although the initial results of phage therapy were promising (3, 4), poorly controlled trials and inconsistent results generated controversy within the scientific community about the efficacy and reproducibility of using phages to treat bacterial infections (5–7). The discovery of penicillin in 1928 and the subsequent arrival of the antibiotic era further cast a shadow on phage therapy (5, 6). As a result, phage therapy was discontinued in Western countries, even as its use continued in Eastern Europe and the former Soviet Union (8–10).
Despite the important success of antibiotics in improving human health, antibiotic resistance has emerged with steadily increasing frequency, rendering many antibiotics ineffective (11–14). Multidrug-resistant bacteria currently constitute one of the most widespread global public health concerns (15–17). More than 2 million people are sickened every year in the United States alone as a result of antibiotic-resistant infections, resulting in at least 23,000 deaths per year (16). The rising tide of antibiotic resistance coupled with the low rate of antibiotic discovery (17, 18) has revived interest in phages as antibacterial agents (19–21).
Unlike most antibiotics, phages are typically highly specific for a particular set of bacterial species or strains and are thus expected to have fewer off-target effects on commensal microflora than antibiotics do (22). The self-replicating nature of phages and the availability of simple, rapid, and low-cost phage production processes are additional advantages for their use as antimicrobials (22). Phages have been used not only to treat and prevent human bacterial infections (9, 23–25) but also to control plant diseases (26–29), detect pathogens (30–33), and assess food safety (34–37).
READ/DOWNLOAD THE ARTICLE
REVIEW ARTICLE
Microbiol., 03 May 2019 | https://doi.org/10.3389/fmicb.2019.00954
Genetic Engineering of Bacteriophages Against Infectious Diseases
 Yibao Chen1,2,
Yibao Chen1,2,  Himanshu Batra3,
Himanshu Batra3,  Junhua Dong1,2,
Junhua Dong1,2,  Cen Chen1,2,
Cen Chen1,2,  Venigalla B. Rao3 and
Venigalla B. Rao3 and  Pan Tao1,2,3*
Pan Tao1,2,3*
Bacteriophages (phages) are the most abundant and widely distributed organisms on Earth, constituting a virtually unlimited resource to explore the development of biomedical therapies. The therapeutic use of phages to treat bacterial infections (“phage therapy”) was conceived by Felix d’Herelle nearly a century ago. However, its power has been realized only recently, largely due to the emergence of multi-antibiotic resistant bacterial pathogens. Progress in technologies, such as high-throughput sequencing, genome editing, and synthetic biology, further opened doors to explore this vast treasure trove. Here, we review some of the emerging themes on the use of phages against infectious diseases. In addition to phage therapy, phages have also been developed as vaccine platforms to deliver antigens as part of virus-like nanoparticles that can stimulate immune responses and prevent pathogen infections. Phage engineering promises to generate phage variants with unique properties for prophylactic and therapeutic applications. These approaches have created momentum to accelerate basic as well as translational phage research and potential development of therapeutics in the near future.
Introduction
Bacteriophages (phages), discovered in the early 20th century independently by Frederick Twort and Felix d’Herelle, are the most abundant organisms on earth with up to 2.5 × 108 phages per ml in natural waters (Bergh et al., 1989). It is well accepted that phages specifically infect bacteria and, therefore, were considered for the development of natural approaches to treat bacterial infections since their discovery (Wittebole et al., 2014; Salmond and Fineran, 2015). However, due to the discovery of antibiotics that provided greater breadth and potency, phage therapy lagged behind although research continued in some Eastern European countries (Chanishvili, 2012, 2016; Wittebole et al., 2014). Therefore, in the following several decades, phages were mainly used as model organisms to explore the basic mechanisms of life and led to the birth of modern molecular biology. One classical example is the demonstration of a central biological question in the early 20th century, the nature of a gene, by “Hershey-Chase experiment” (also called “Waring blender experiment”) (Salmond and Fineran, 2015). This elegant experiment demonstrated that DNA, not protein, is the genetic material of T2 phage.
Recently, the emergence of multi-antibiotic resistant bacterial pathogens and the low rate of new antibiotic discovery brought new urgency to develop phage-based therapies (Lu and Koeris, 2011; Viertel et al., 2014; Domingo-Calap and Delgado-Martinez, 2018). A striking example is the recent San Diego patient who was infected by multi-drug resistant Acinetobacter baumannii stain during travelling to Egypt. The patient went into a coma for nearly 2 months but awoke 2 days after intravenous injection of a phage cocktail that lyses this bacterium and finally completely recovered (Schooley et al., 2017). With recent advances, particularly the genome engineering (Martel and Moineau, 2014; Ando et al., 2015; Lemay et al., 2017; Tao et al., 2017b; Kilcher et al., 2018), the applications of phages have greatly expanded. In addition to its use in antibacterial therapy, phages were used in synthetic biology (Lemire et al., 2018), material science (Cao et al., 2016), and biomedical fields (Cao et al., 2018; Tao et al., 2018c). Considering the abundance and diversity, there is vast potential to engineer phages for different applications. In this review, we will focus on the applications of phages in infectious disease, in particular, vaccine development and phage therapy. We will discuss the phage engineering strategies and how these can equip the phages with the ability to advance the vaccine and phage therapy fields.
REVIEW ARTICLE
Microbiol., 03 May 2019 | https://doi.org/10.3389/fmicb.2019.00954
Genetic Engineering of Bacteriophages Against Infectious Diseases
 Yibao Chen1,2,
Yibao Chen1,2,  Himanshu Batra3,
Himanshu Batra3,  Junhua Dong1,2,
Junhua Dong1,2,  Cen Chen1,2,
Cen Chen1,2,  Venigalla B. Rao3 and
Venigalla B. Rao3 and  Pan Tao1,2,3*
Pan Tao1,2,3*Bacteriophages (phages) are the most abundant and widely distributed organisms on Earth, constituting a virtually unlimited resource to explore the development of biomedical therapies. The therapeutic use of phages to treat bacterial infections (“phage therapy”) was conceived by Felix d’Herelle nearly a century ago. However, its power has been realized only recently, largely due to the emergence of multi-antibiotic resistant bacterial pathogens. Progress in technologies, such as high-throughput sequencing, genome editing, and synthetic biology, further opened doors to explore this vast treasure trove. Here, we review some of the emerging themes on the use of phages against infectious diseases. In addition to phage therapy, phages have also been developed as vaccine platforms to deliver antigens as part of virus-like nanoparticles that can stimulate immune responses and prevent pathogen infections. Phage engineering promises to generate phage variants with unique properties for prophylactic and therapeutic applications. These approaches have created momentum to accelerate basic as well as translational phage research and potential development of therapeutics in the near future.
Introduction
Bacteriophages (phages), discovered in the early 20th century independently by Frederick Twort and Felix d’Herelle, are the most abundant organisms on earth with up to 2.5 × 108 phages per ml in natural waters (Bergh et al., 1989). It is well accepted that phages specifically infect bacteria and, therefore, were considered for the development of natural approaches to treat bacterial infections since their discovery (Wittebole et al., 2014; Salmond and Fineran, 2015). However, due to the discovery of antibiotics that provided greater breadth and potency, phage therapy lagged behind although research continued in some Eastern European countries (Chanishvili, 2012, 2016; Wittebole et al., 2014). Therefore, in the following several decades, phages were mainly used as model organisms to explore the basic mechanisms of life and led to the birth of modern molecular biology. One classical example is the demonstration of a central biological question in the early 20th century, the nature of a gene, by “Hershey-Chase experiment” (also called “Waring blender experiment”) (Salmond and Fineran, 2015). This elegant experiment demonstrated that DNA, not protein, is the genetic material of T2 phage.
Recently, the emergence of multi-antibiotic resistant bacterial pathogens and the low rate of new antibiotic discovery brought new urgency to develop phage-based therapies (Lu and Koeris, 2011; Viertel et al., 2014; Domingo-Calap and Delgado-Martinez, 2018). A striking example is the recent San Diego patient who was infected by multi-drug resistant Acinetobacter baumannii stain during travelling to Egypt. The patient went into a coma for nearly 2 months but awoke 2 days after intravenous injection of a phage cocktail that lyses this bacterium and finally completely recovered (Schooley et al., 2017). With recent advances, particularly the genome engineering (Martel and Moineau, 2014; Ando et al., 2015; Lemay et al., 2017; Tao et al., 2017b; Kilcher et al., 2018), the applications of phages have greatly expanded. In addition to its use in antibacterial therapy, phages were used in synthetic biology (Lemire et al., 2018), material science (Cao et al., 2016), and biomedical fields (Cao et al., 2018; Tao et al., 2018c). Considering the abundance and diversity, there is vast potential to engineer phages for different applications. In this review, we will focus on the applications of phages in infectious disease, in particular, vaccine development and phage therapy. We will discuss the phage engineering strategies and how these can equip the phages with the ability to advance the vaccine and phage therapy fields.
1College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
2The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan, China
3Department of Biology, The Catholic University of America, Washington, DC, United States
2The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan, China
3Department of Biology, The Catholic University of America, Washington, DC, United States
READ / DOWNLOAD THE REST OF THE ARTICLE HERE
Allosteric inhibition of CRISPR-Cas9 by bacteriophage-derived peptides
Yan-ru Cui,
Shao-jie Wang,
Jun Chen,
Jie Li,
Wenzhang Chen,
Shuyue Wang,
Bing Meng,
Wei Zhu,
Zhuhong Zhang,
Bei Yang,
Biao Jiang,
Guang Yang,
Peixiang Ma &
Jia Liu
Genome Biology volume 21, Article number: 51 (2020) Cite this article
Research
Open Access
Published: 26 February 2020
Abstract
Background
CRISPR-Cas9 has been developed as a therapeutic agent for various infectious and genetic diseases. In many clinically relevant applications, constitutively active CRISPR-Cas9 is delivered into human cells without a temporal control system. Excessive and prolonged expression of CRISPR-Cas9 can lead to elevated off-target cleavage. The need for modulating CRISPR-Cas9 activity over time and dose has created the demand of developing CRISPR-Cas off switches. Protein and small molecule-based CRISPR-Cas inhibitors have been reported in previous studies.
Allosteric inhibition of CRISPR-Cas9 by bacteriophage-derived peptides
Yan-ru Cui,
Shao-jie Wang,
Jun Chen,
Jie Li,
Wenzhang Chen,
Shuyue Wang,
Bing Meng,
Wei Zhu,
Zhuhong Zhang,
Bei Yang,
Biao Jiang,
Guang Yang,
Peixiang Ma &
Jia Liu
Genome Biology volume 21, Article number: 51 (2020) Cite this article
Research
Open Access
Published: 26 February 2020
Abstract
Background
CRISPR-Cas9 has been developed as a therapeutic agent for various infectious and genetic diseases. In many clinically relevant applications, constitutively active CRISPR-Cas9 is delivered into human cells without a temporal control system. Excessive and prolonged expression of CRISPR-Cas9 can lead to elevated off-target cleavage. The need for modulating CRISPR-Cas9 activity over time and dose has created the demand of developing CRISPR-Cas off switches. Protein and small molecule-based CRISPR-Cas inhibitors have been reported in previous studies.
ResultsWe report the discovery of Cas9-inhibiting peptides from inoviridae bacteriophages. These peptides, derived from the periplasmic domain of phage major coat protein G8P (G8PPD), can inhibit the in vitro activity of Streptococcus pyogenes Cas9 (SpCas9) proteins in an allosteric manner. Importantly, the inhibitory activity of G8PPD on SpCas9 is dependent on the order of guide RNA addition. Ectopic expression of full-length G8P (G8PFL) or G8PPD in human cells can inactivate the genome-editing activity of SpyCas9 with minimum alterations of the mutation patterns. Furthermore, unlike the anti-CRISPR protein AcrII4A that completely abolishes the cellular activity of CRISPR-Cas9, G8P co-transfection can reduce the off-target activity of co-transfected SpCas9 while retaining its on-target activity.
Conclusion
G8Ps discovered in the current study represent the first anti-CRISPR peptides that can allosterically inactivate CRISPR-Cas9. This finding may provide insights into developing next-generation CRISPR-Cas inhibitors for precision genome engineering.
G8Ps discovered in the current study represent the first anti-CRISPR peptides that can allosterically inactivate CRISPR-Cas9. This finding may provide insights into developing next-generation CRISPR-Cas inhibitors for precision genome engineering.
READ/DOWNLOAD HERE
Heterogeneous Diversity of Spacers within CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)
Article (PDF Available) in Physical Review Letters 105(12):128102 · September 2010 with 231
Download full-text PDF
DOI: 10.1103/PhysRevLett.105.128102 · Source: PubMedCite this publication

Jiankui he
Southern University of Science and Technology

Michael Deem
Rice University
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) in bacterial and archaeal DNA have recently been shown to be a new type of antiviral immune system in these organisms. We here study the diversity of spacers in CRISPR under selective pressure. We propose a population dynamics model that explains the biological observation that the leader-proximal end of CRISPR is more diversified and the leader-distal end of CRISPR is more conserved. This result is shown to be in agreement with recent experiments. Our results show that the CRISPR spacer structure is influenced by and provides a record of the viral challenges that bacteria face.
https://www.researchgate.net/publication/46424214_Heterogeneous_Diversity_of_Spacers_within_CRISPR_Clustered_Regularly_Interspaced_Short_Palindromic_Repeats
Heterogeneous Diversity of Spacers within CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)
Article (PDF Available) in Physical Review Letters 105(12):128102 · September 2010 with 231
Download full-text PDF
DOI: 10.1103/PhysRevLett.105.128102 · Source: PubMedCite this publication

Jiankui he
Southern University of Science and Technology

Michael Deem
Rice University
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) in bacterial and archaeal DNA have recently been shown to be a new type of antiviral immune system in these organisms. We here study the diversity of spacers in CRISPR under selective pressure. We propose a population dynamics model that explains the biological observation that the leader-proximal end of CRISPR is more diversified and the leader-distal end of CRISPR is more conserved. This result is shown to be in agreement with recent experiments. Our results show that the CRISPR spacer structure is influenced by and provides a record of the viral challenges that bacteria face.
https://www.researchgate.net/publication/46424214_Heterogeneous_Diversity_of_Spacers_within_CRISPR_Clustered_Regularly_Interspaced_Short_Palindromic_Repeats

Volume 366
Issue 9
May 2019
Article Contents
ABSTRACT
INTRODUCTION
BIOLOGICAL RELEVANCE OF ANTI-CRISPR PROTEINS
MECHANISMS AND STRUCTURES OF ANTI-CRISPR PROTEINS
APPLICATIONS OF ANTI-CRISPR PROTEINS
OUTLOOK
FUNDING
REFERENCES
MINI REVIEW
Keeping CRISPR in check: diverse mechanisms of phage-encoded anti-CRISPRS
Despoina Trasanidou, Ana Sousa Gerós, Prarthana Mohanraju, Anna Cornelia Nieuwenweg, Franklin L Nobrega, Raymond H J Staals
FEMS Microbiology Letters, Volume 366, Issue 9, May 2019, fnz098, https://doi.org/10.1093/femsle/fnz098
Published: 11 May 2019
ABSTRACT
CRISPR-Cas represents the only adaptive immune system of prokaryotes known to date. These immune systems are widespread among bacteria and archaea, and provide protection against invasion of mobile genetic elements, such as bacteriophages and plasmids. As a result of the arms-race between phages and their prokaryotic hosts, phages have evolved inhibitors known as anti-CRISPR (Acr) proteins to evade CRISPR immunity. In the recent years, several Acr proteins have been described in both temperate and virulent phages targeting diverse CRISPR-Cas systems. Here, we describe the strategies of Acr discovery and the multiple molecular mechanisms by which these proteins operate to inhibit CRISPR immunity. We discuss the biological relevance of Acr proteins and speculate on the implications of their activity for the development of improved CRISPR-based research and biotechnological tools.

The physicist's guide to one of biotechnology's hottest new topics: CRISPR-Cas
Melia E Bonomo1,3 and Michael W Deem1,2,3,4
Published 30 April 2018 • © 2018 IOP Publishing Ltd
Physical Biology, Volume 15, Number 4
DownloadArticle PDF
Article information
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) constitute a multi-functional, constantly evolving immune system in bacteria and archaea cells. A heritable, molecular memory is generated of phage, plasmids, or other mobile genetic elements that attempt to attack the cell. This memory is used to recognize and interfere with subsequent invasions from the same genetic elements. This versatile prokaryotic tool has also been used to advance applications in biotechnology. Here we review a large body of CRISPR-Cas research to explore themes of evolution and selection, population dynamics, horizontal gene transfer, specific and cross-reactive interactions, cost and regulation, non-immunological CRISPR functions that boost host cell robustness, as well as applicable mechanisms for efficient and specific genetic engineering. We offer future directions that can be addressed by the physics community. Physical understanding of the CRISPR-Cas system will advance uses in biotechnology, such as developing cell lines and animal models, cell labeling and information storage, combatting antibiotic resistance, and human therapeutics.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 3.0 licence. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
In 1987, Ishino and colleagues had set out to identify the encoded protein and primary structure of a particular gene in Escherichia coli by analyzing its chromosomal DNA segment and flanking regions [1]. They found an interesting sequence structure at the gene's 3'-end flanking region, in which five homologous sequences of 29 nucleotides were arranged as direct repeats with 32-nucleotide sequences spaced between them. Little did they know that their discovery would prove to have critical immunological significance. It was not until 2000 that these mysterious repeated genomic elements were revisited when Mojica and colleagues searched the available microbial genome database and found many organisms that contained partially palindromic sequences of 24–40 basepairs with 20–58 basepair sequences spaced between them [2]. These were found in almost all archaea, about half of bacteria, no viruses, and no eukaryotes. Related and unrelated species had nearly identical structure in these repeat sequence units. The sequences in between, called 'spacers', were unique to an individual locus and were not found in other genomes [3]. After many suggested abbreviations, including SRSRs, short regularly spaced repeats, and SPIDR, spacers interspersed direct repeats, the scientific community settled on calling these elements clustered regularly interspaced short palindromic repeats, or CRISPR.
SEE
https://plawiuk.blogspot.com/search?q=BACTERIOPHAGES
https://plawiuk.blogspot.com/search?q=PHAGES





