Remembering the Work of G.N. Ramachandran and Others in the Time of COVID-19
29/08/2020

G.N. Ramachandran. Photo: Current Science, vol. 80, no. 8, April 2001/fair use.
The whole world is reeling under the impact of the COVID-19 pandemic. The safest way out depends on manufacturing an effective vaccine. Many vaccine candidates are currently in different stages of clinical trials. A quick scan of the list suggests over half the candidates are attempting to use the novel coronavirus’s spike protein to generate immunity, so getting its structure right is critical.
G.N. Ramachandran’s work from the 1960s is notable in this regard. I was first introduced to his influence on structural biology as a graduate student and have always wished he were better known outside this niche field. His work has guided the design of protein-based vaccines and therapeutics touching everyday life. However, his isn’t a household name even in India.
Ramachandran began his academic career as a student of electrical engineering at the Indian Institute of Science, Bengaluru. He was soon pulled into the physics department by C.V. Raman, who said, “I am admitting Ramachandran into my department as he is a bit too bright to be in yours.”
Under Raman, Ramachandran began research into the field of optics and X-ray topography. After graduation, he earned a PhD at the University of Cambridge, in the laboratory of Lawrence Bragg. He focused on X-ray crystallography, a technique used to analyse the structures of proteins.
Around the same time, the American physicist Linus Pauling was conducting groundbreaking work on the nature and structure of proteins. Pauling’s teaching and writings had a great influence on Ramachandran. So when the latter became a professor of physics at the University of Madras in 1952, he set up an X-ray crystallography laboratory to study biological structures.
Proteins are made of long-chain polymers called polypeptides. There can be one or multiple polypeptide chains that come together to form a protein. The novel coronavirus’s spike protein comprises three identical peptide chains. Each chain consists of repeating units called amino acids. The chain folds into intricate shapes or motifs, such as helices and sheets. These shapes are significant; misfolded proteins often give rise to debilitating diseases
.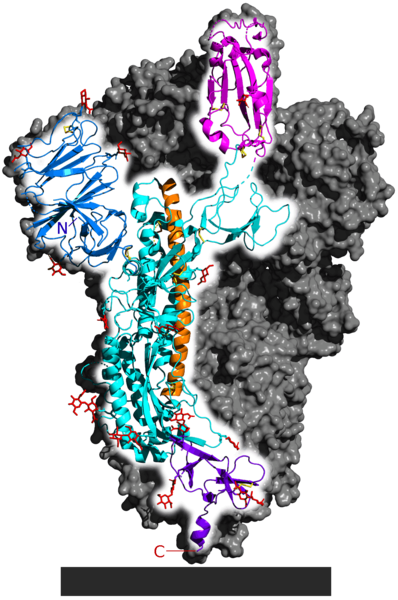

Spike glycoprotein from SARS-CoV-2. Image: 5-HT2AR/Wikimedia CommonsPIN IT
The long orange region in the spike protein is the alpha helix and the pink region is a beta sheet. Mixing and matching of motifs form different protein structures required to ensure the protein performs its function. In this case, the spike protein’s overall structure helps to bind with a protein on the surface of cells in the human nose and mouth.
The order of amino acids within a peptide chain dictates how it will fold. Amino acids take their name from the nitrogen-containing molecule (amino) on one end and the carbonyl group (acid) on the other, with a connecting carbon atom in the middle (Cα). The differences between amino acids come from a third part, called the side chain (R), which attaches to the central carbon atom
The long orange region in the spike protein is the alpha helix and the pink region is a beta sheet. Mixing and matching of motifs form different protein structures required to ensure the protein performs its function. In this case, the spike protein’s overall structure helps to bind with a protein on the surface of cells in the human nose and mouth.
The order of amino acids within a peptide chain dictates how it will fold. Amino acids take their name from the nitrogen-containing molecule (amino) on one end and the carbonyl group (acid) on the other, with a connecting carbon atom in the middle (Cα). The differences between amino acids come from a third part, called the side chain (R), which attaches to the central carbon atom
.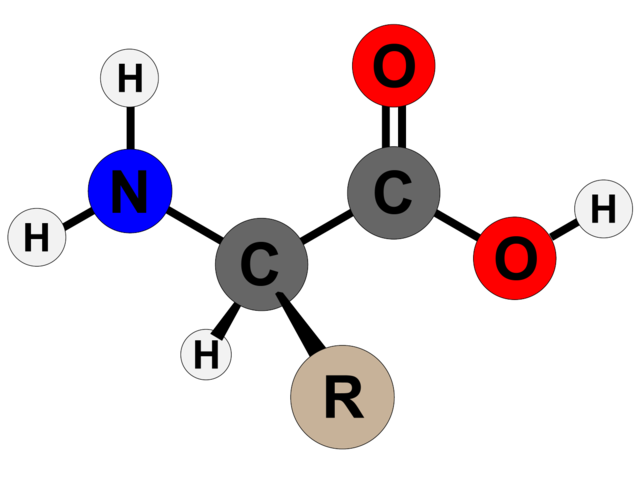

The structure of an un-ionised amino acid. Image: Techguy78/Wikimedia Commons, CC BY-SA 4.0PIN IT
In the mid-20th century, the idea of firing beams of X-rays at molecules to elucidate their structure based on how they diffracted the beams was new. In 1951, Pauling, Robert Corey and Herman Branson published their descriptions of the alpha helix and beta sheet motifs. Ramachandran wanted to continue this work, and chose to study the structure of collagen first.
Collagen is protein found abundantly in the human body: it makes up the bulk of our skin, cartilage and connective tissues. Together with his postdoctoral fellow, Gopinath Kartha, Ramachandran proposed a triple helix structure for collagen – also called the Madras helix. However, Alexander Rich and Francis Crick contested this idea because they thought this structure allowed little space between atoms than was compatible with the prevailing understanding of chemistry.
This set Ramachandran and his colleagues C. Ramakrishnan and V. Sasisekharan on the path to further understand and describe the structures of polypeptide chains. They conducted a survey of the crystal structures available to determine how close two atoms could approach, and thus deduce the permissible interatomic distances within a polypeptide chain.
They also found that in a chain, each amino acid can only rotate around two bonds. Based on this they characterised two angles – phi (φ), the angle between the nitrogen atom and the central carbon atom, and psi (ψ), the angle between the carbonyl carbon atom and the central carbon. The figure below shows the two angles
In the mid-20th century, the idea of firing beams of X-rays at molecules to elucidate their structure based on how they diffracted the beams was new. In 1951, Pauling, Robert Corey and Herman Branson published their descriptions of the alpha helix and beta sheet motifs. Ramachandran wanted to continue this work, and chose to study the structure of collagen first.
Collagen is protein found abundantly in the human body: it makes up the bulk of our skin, cartilage and connective tissues. Together with his postdoctoral fellow, Gopinath Kartha, Ramachandran proposed a triple helix structure for collagen – also called the Madras helix. However, Alexander Rich and Francis Crick contested this idea because they thought this structure allowed little space between atoms than was compatible with the prevailing understanding of chemistry.
This set Ramachandran and his colleagues C. Ramakrishnan and V. Sasisekharan on the path to further understand and describe the structures of polypeptide chains. They conducted a survey of the crystal structures available to determine how close two atoms could approach, and thus deduce the permissible interatomic distances within a polypeptide chain.
They also found that in a chain, each amino acid can only rotate around two bonds. Based on this they characterised two angles – phi (φ), the angle between the nitrogen atom and the central carbon atom, and psi (ψ), the angle between the carbonyl carbon atom and the central carbon. The figure below shows the two angles
.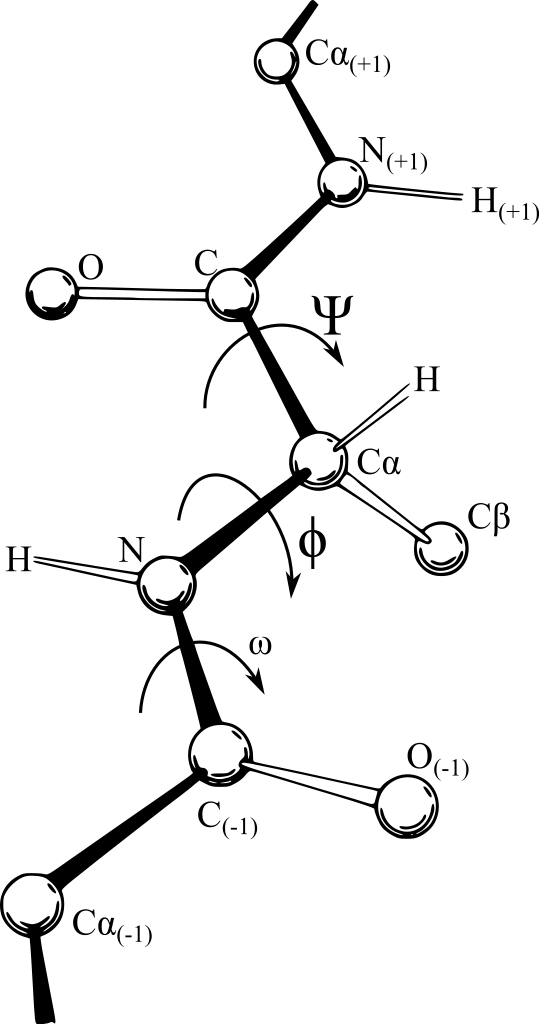

A protein backbone showing the phi and psi angles.
Image: Dcrjsr/Wikimedia Commons, CC BY 3.0 PIN IT
Now, they could plot all possible combinations of these angles within a given polypeptide sequence against each other, and eliminate any combination that violated the interatomic contact limits. And voila! They managed to reduce a messy biological problem to simple considerations in elementary mathematics.
The resulting Ramachandran plot changed how biochemists studied molecules of interest and unraveled complicated biological processes. It’s a plot with phi on one axis and psi on the other. At a glance, the graph revealed islands of possible combinations separated by seas of impossible structures. If a molecule’s angles pulled it into the sea, biochemists would know it couldn’t exist in the body and follow the laws of physical chemistry at the same time
Now, they could plot all possible combinations of these angles within a given polypeptide sequence against each other, and eliminate any combination that violated the interatomic contact limits. And voila! They managed to reduce a messy biological problem to simple considerations in elementary mathematics.
The resulting Ramachandran plot changed how biochemists studied molecules of interest and unraveled complicated biological processes. It’s a plot with phi on one axis and psi on the other. At a glance, the graph revealed islands of possible combinations separated by seas of impossible structures. If a molecule’s angles pulled it into the sea, biochemists would know it couldn’t exist in the body and follow the laws of physical chemistry at the same time
.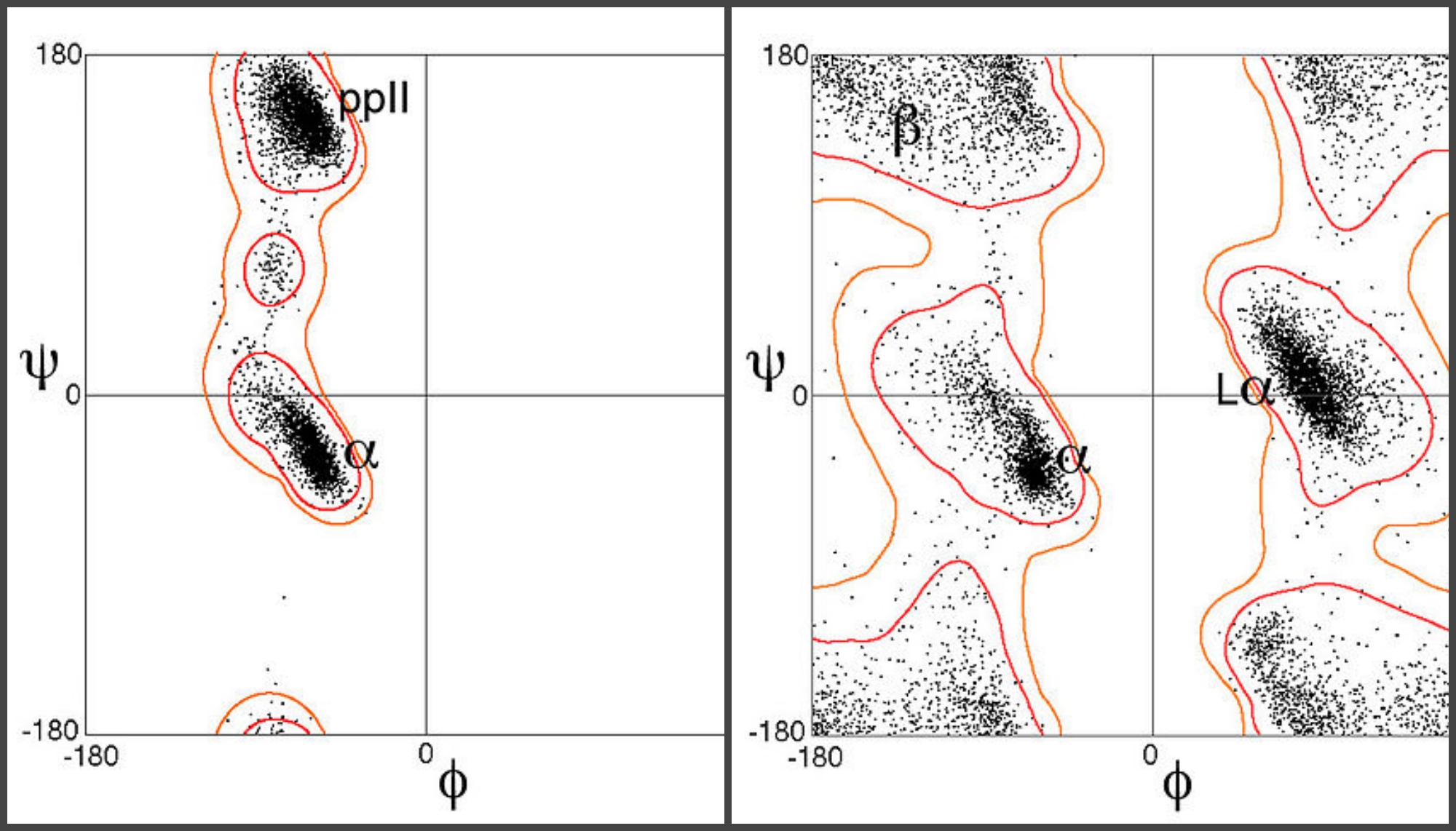

Ramachandran plots for two amino acids, proline (left) and glycine (right). Images: Dcrjsr/Wikimedia Commons, CC BY 3.0PIN IT
More broadly, biochemists today can quickly understand which structures are possible and which aren’t, and compare known and unknown structures in an intuitive manner. The Ramachandran plot has also become an elegant way to introduce students to protein structural biology and help them understand how structure and function are related to each other.
When Ramachandran died in 2001, at the age of 78, the tributes flooded in. One in particular, by Janet Thornton, published in an obituary by Easwara Subramanian, went thus:
“I have never met Professor Ramachandran, but his contribution … ranks with Pauling’s discovery of the α-helix. It never fails to excite me, when I see the Ramachandran plot and realise how much of the beauty and order of protein structures is encapsulated by this plot. I also think that this major discovery highlights the importance of clear thought and vision that do not always need expensive equipment and huge teams of people.”
The plot’s remarkable longevity is unsurprising given its endless significance in protein structural biology. It is vital that researchers independently validate different biological structures so that others who use those biological objects, like proteins, to develop drugs and vaccines can be sure they know the structure well enough. There exist a variety of algorithms that can check a structure’s accuracy using updated Ramachandran plots.
When they need to check the structure of an unknown protein, researchers compare data obtained from crystallographic studies and from theoretical calculations. The more the plots match, the more confident scientists can be that their structure is the right one.
Ramachandran’s life is a testament to the power of curiosity, intellect and determination. Science education can be biased in favour of assigning credit to individuals, often white men of the West, over groups. In fact, science is an inherently collaborative endeavour, with each generation building on the work of previous ones. So just as we celebrate Ramachandran’s life and work, let’s also celebrate the lives and work of his students and collaborators – who together helped advance the science of protein structural biology such that it has risen to the occasion in our present crisis as well.
Deepika Calidas is a biochemist. Her last position was as a postdoctoral fellow at the Johns Hopkins School of Medicine.
More broadly, biochemists today can quickly understand which structures are possible and which aren’t, and compare known and unknown structures in an intuitive manner. The Ramachandran plot has also become an elegant way to introduce students to protein structural biology and help them understand how structure and function are related to each other.
When Ramachandran died in 2001, at the age of 78, the tributes flooded in. One in particular, by Janet Thornton, published in an obituary by Easwara Subramanian, went thus:
“I have never met Professor Ramachandran, but his contribution … ranks with Pauling’s discovery of the α-helix. It never fails to excite me, when I see the Ramachandran plot and realise how much of the beauty and order of protein structures is encapsulated by this plot. I also think that this major discovery highlights the importance of clear thought and vision that do not always need expensive equipment and huge teams of people.”
The plot’s remarkable longevity is unsurprising given its endless significance in protein structural biology. It is vital that researchers independently validate different biological structures so that others who use those biological objects, like proteins, to develop drugs and vaccines can be sure they know the structure well enough. There exist a variety of algorithms that can check a structure’s accuracy using updated Ramachandran plots.
When they need to check the structure of an unknown protein, researchers compare data obtained from crystallographic studies and from theoretical calculations. The more the plots match, the more confident scientists can be that their structure is the right one.
Ramachandran’s life is a testament to the power of curiosity, intellect and determination. Science education can be biased in favour of assigning credit to individuals, often white men of the West, over groups. In fact, science is an inherently collaborative endeavour, with each generation building on the work of previous ones. So just as we celebrate Ramachandran’s life and work, let’s also celebrate the lives and work of his students and collaborators – who together helped advance the science of protein structural biology such that it has risen to the occasion in our present crisis as well.
Deepika Calidas is a biochemist. Her last position was as a postdoctoral fellow at the Johns Hopkins School of Medicine.
No comments:
Post a Comment