SHANGRILA
Expedition finds Tibetan lakes harbor bacteria that produce antibiotics
Skoltech researchers and their colleagues from China and Russia have discovered that the waters and soils of the Tibetan Plateau are teeming with bacteria that produce antibiotics. While none of the antimicrobial compounds identified by the team are new to science, the findings bring certain hope amid the outbreak of bacterial resistance triggered by irresponsible drug use.
Further investigation of remote and hard-to-reach habitats in Tibet and elsewhere could eventually uncover new antibiotics. The team's findings are reported in Frontiers in Microbiology and Microorganisms.
Every time someone takes antibiotics, microbes get a glimpse of humanity's ultimate secret weapon against bacterial infections. The few surviving bacteria hand down the protective mechanisms they happened to have to their offspring. This wouldn't matter nearly as much if people only used antibiotics on prescription, but as it stands, many of our most potent antimicrobials are not so secret anymore: Bacteria are growing resistant to them.
The biomedical community responds to bacterial resistance by promoting responsible drug use and searching for new drugs.
"There are two basic approaches to discovering antibiotics," says Assistant Professor Dmitrii Lukianov from Skoltech Bio, who co-authored both studies. "One thing you can do is browse chemical libraries—vast databases containing hundreds of thousands of compounds, which you can test for antibiotic activity. That search can be facilitated by machine learning algorithms. Alternatively, you can look for drug candidates in nature—for example in soil—because bacteria dwelling there use antibiotics to fight other bacteria."
The samples first went to Beijing Key Laboratory of Antimicrobial Agents. There, the researchers cultivated the samples in a nutrient-rich medium known as culture broth. The liquid cultures obtained by the team were then tested for antimicrobial activity. The bacterial strains found to produce antibiotic compounds underwent genetic analysis. The researchers then pitted the analyzed strains against clinically important bacterial strains known to cause disease in humans.
The next stage is where the Skoltech laboratory took over. After receiving the dry extracts of the cultural liquids containing antibiotic compounds, Lukianov and his colleagues set out to determine the mechanisms of antimicrobial action involved using a so-called reporter system, developed by study co-author and Bio Center Professor Petr Sergiev, among other researchers.
"There are several ways antibiotics can harm bacteria: by affecting protein synthesis, DNA replication, RNA transcription, cell wall synthesis, or key metabolic processes," Lukianov explains.
"Skoltech and MSU scientists created a reporter system that enables us to distinguish compounds with two mechanisms of action. First, antibiotics that suppress the production of protein by bacteria. Second, compounds that affect DNA replication or RNA transcription." All the other mechanisms of action were classified as the third group.
To determine which mechanism was at play in any given case, the system relies on a double "reporter strain" of the E. coli bacterium. It's a laboratory strain artificially devoid of certain defense mechanisms, making the effects of antibiotics easier to discern and measure.
To run the test, the scientists cultivated the reporter strain on petri dishes and subjected it to the Tibetan cultural liquid extracts, as well as to controls—standard antibiotics that represent each of the two main mechanisms. Based on how deadly the analyzed compounds proved to the reporter strain, when compared with commonly used antibiotics, the team could determine which mechanism was involved.
The researchers then used two highly precise analytical techniques for identifying compounds—chromatography and mass spectrometry—to tell which chemical species were contained in the extracts. Among them were hedamycins and kidamycins, in saline lake water, as well as rifamycins from the Tibetan soil samples.
"While compounds from these families are known antibiotics, their presence in such remote regions and at such an elevation above sea level is not something you would readily suspect," Lukianov comments. "There might be other, unfamiliar antibiotics lurking out there, too. And perhaps next time we'll be lucky enough to find them."
However, if you decide to dig for revolutionary new antibiotics in your backyard, chances are you are going to discover compounds long known to science and therefore of limited interest to the medical community. Not so for remote areas that are sparsely populated and hard to access. Such as Tibet.
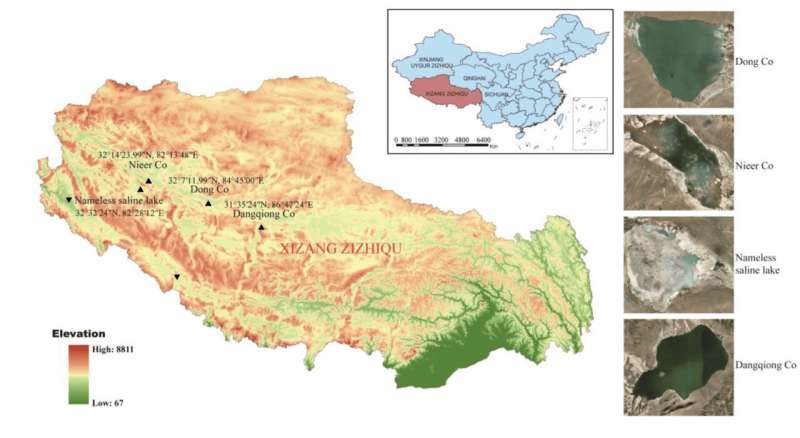
The Chinese co-authors of the two studies reported in this story went for an expedition to the Tibetan Plateau and collected water and soil samples from a number of locations across the highlands, including four saline lakes.
More information: Lifang Liu et al, Bioprospecting for the soil-derived actinobacteria and bioactive secondary metabolites on the Western Qinghai-Tibet Plateau, Frontiers in Microbiology (2023). DOI: 10.3389/fmicb.2023.1247001
Shao-Wei Liu et al, Bioprospecting of Actinobacterial Diversity and Antibacterial Secondary Metabolites from the Sediments of Four Saline Lakes on the Northern Tibetan Plateau, Microorganisms (2023). DOI: 10.3390/microorganisms11102475
No comments:
Post a Comment