Contamination analysis of Arctic ice samples as planetary field analogs and implications for future life-detection missions to Europa and Enceladus
Scientific Reports 12, Article number: 12379 (2022)
- Open Access
- Published:
Abstract
Missions to detect extraterrestrial life are being designed to visit Europa and Enceladus in the next decades. The contact between the mission payload and the habitable subsurface of these satellites involves significant risk of forward contamination. The standardization of protocols to decontaminate ice cores from planetary field analogs of icy moons, and monitor the contamination in downstream analysis, has a direct application for developing clean approaches crucial to life detection missions in these satellites. Here we developed a comprehensive protocol that can be used to monitor and minimize the contamination of Arctic ice cores in processing and downstream analysis. We physically removed the exterior layers of ice cores to minimize bioburden from sampling. To monitor contamination, we constructed artificial controls and applied culture-dependent and culture-independent techniques such as 16S rRNA amplicon sequencing. We identified 13 bacterial contaminants, including a radioresistant species. This protocol decreases the contamination risk, provides quantitative and qualitative information about contamination agents, and allows validation of the results obtained. This study highlights the importance of decreasing and evaluating prokaryotic contamination in the processing of polar ice cores, including in their use as analogs of Europa and Enceladus.
Introduction
Europa and Enceladus are two icy moons from our solar system identified as ocean worlds due to the presence of a liquid ocean under their icy surface1,2. Europa has also secondary liquid water reservoirs, perched in the ice and closer to the active surface3. The water bodies present in these satellites are considered habitable environments4. Several concepts for future lander missions to these moons have been developed; e.g., Europa Lander, Enceladus Orbilander, and Joint Europa Mission (JEM)5,6,7. These missions will need to drill and sample a layer of ice (exact extension still undetermined) to eventually reach interface water.
The Committee of Space Research (COSPAR) recommends that the study of methods of bioburden reduction for these missions should reflect the type of environments found on Europa or Enceladus, focusing on Earth organisms most likely to survive on these moons, such as cold and radiation tolerant organisms8,9,10. Environments on Earth that exhibit similar extreme conditions as planets and moons in our solar system are called planetary field analogues11,12,13. Both the Arctic and Antarctic offer locations that mimic environments present in the icy moons of Jupiter and Saturn12. These locations are populated by extremophiles—organisms adapted to survive these severe conditions such as extreme temperature and pH, dryness, oxidation, UV radiation, high pressure, and high salinity14,15. Microbes from these habitats are viable despite hundreds to thousands of years in terrestrial glaciers and cryo-permafrost environments16,17, increasing the plausibility of finding putative life forms in Europa and Enceladus. Polar extremophiles are also known to survive sterilization procedures for planetary protection18 and under space conditions on-board the International Space Station (ISS)19.
The challenges of sampling, processing, and analyzing Arctic and Antarctic ice samples are the closest to those of future life-detection missions in these satellites20,21. The constraints include difficulties in sampling, analysis of low biomass samples, and the need to minimize and monitor potential sources22 of contamination from mesophilic environments on Earth, where microbes are ubiquitous and in high abundance.
When studying recent terrestrial ice, contamination is critical, and sources are mainly due to equipment such as ice corers, handling, and transportation20,23,24. In the context of planetary field analogs, we add more contamination sources such as snow, air, and soil microbes that are part of the atmospheric and soil microbiome but are not expected to be present in icy worlds with thin atmospheres and no regolith25. Studies from the last 20 years of ice core research mention the use of sterile equipment in the field while drilling the ice cores (Table S1—Supplementary material). Codes of conduct and clean protocols to sample pristine subglacial lakes in Antarctica have also been created recently22,24, which represents a challenging and laudable effort by the scientific community to conserve these unique microbial ecosystems. However, the potential sources of contamination are not limited to the field. During manipulations of ice cores in the laboratory, microbial contaminants can be introduced from the laboratory air, equipment, materials, reagents, or even by humans during downstream analysis such as filtration, DNA extraction (the kitome), amplification, and sequencing, or during cultivation in a nutritive medium. These procedures will be robotized in lander space missions; however, they still represent more layers of contact between man-made equipment coming from Earth and extraterrestrial samples. Sterilization methods used for equipment cannot be directly applied on ice samples21. Controlled heat, UV-C, and chemical disinfectants such as ethanol, benzethonium chloride, and sodium hypochlorite have been used directly in the exterior of ice samples being especially efficient in reducing active contaminants in cultivation work20,23,26. However, they are not adequate for life detection, molecular methods of low-biomass ice samples, or the integrity of other microbial analyses. For example, ethanol is an effective disinfectant to decrease contamination for culture-dependent analysis, however, it does not destroy DNA molecules20. While excising the external layers of the ice cores has proved effective in removing most contaminating cells20,23,26,27, without compromising the interior native biota, no known protocol can completely prevent contamination, which leads us to the last possible resource for an ethical sampling and processing methodology: contamination monitoring through the use of background controls27 that have proved to be very effective27,28. Culture-dependent and -independent analyses9,27,29,30,31,32,33,34,35,36,37,38,39 have been used in background controls and in the planetary protection context. In past studies on ice cores, culture-dependent investigations appeared to instigate more care to prevent contaminants in comparison with culture-independent techniques (Table S1—Supplementary material). This is likely because DNA contamination from the laboratory air or sterile material is commonly considered insignificant, due to its presumably low representation in comparison with the microbial load of the whole community present in the samples, which is now overcome by the increased sensitivity of polymerase chain reaction (PCR) and DNA sequencing techniques. The lack of standardized methodology adopted to decrease and monitor contaminations in ice core analysis, similar to what already exists to sampling22,24, remains a limiting issue for the scientific integrity of the acquired data in icy planetary field analogs, as well as for the design of proven and robust protocols for the future lander and return missions to icy moons. As a result, the identity and function of microbial contaminants expected from ice core analysis remains a challenge in the field of planetary protection.
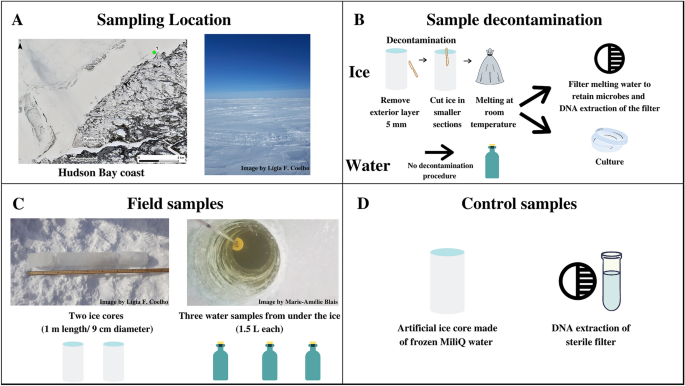
In this study, we propose a multidimensional approach to restrict and monitor the contamination inherent in the processing and analyzing ice samples, combining the most effective methods presented in the last 20 years on ice core studies (Table S1—Supplementary material). The decontamination methodology for the ice core surface was mechanical to decrease contaminants while preserving the natural biota. We constructed background controls (Fig. 1): an artificial ice core made of sterile MilliQ water (processing control) and a DNA extraction control sample (DNA-extraction control). We used both culture-dependent and -independent techniques, such as 16S rRNA gene amplicon sequencing of metagenomic DNA samples, accessing several levels of visible and quantifiable prokaryotic contamination. We identified the contaminating microorganisms of the present study using an established ribosomal marker database to understand the astrobiology relevance of the contaminants. Such a decontamination protocol would be suitable for the design of life-detection experiments on planetary field analogs of Europa and Enceladus, targeting ice cores that may serve as a proxy for habitats of putative extraterrestrial communities. Also, our protocol will serve as a testbed for procedures of decontamination of samples from future landing/return missions to the icy moons.
(A) Sampling location on the east coast of Hudson Bay, Quebec, Canada, latitude 55.39° N; longitude 77.61° W (Map data © Sentinel-2), and the salinity measured on-site (% by mass). (B) Sampling decontamination procedure and following processing preceding culture-dependent and culture-independent analysis. (C) Description of environmental samples with respective replicates: ice cores (duplicate—Ice 1, Ice 2), and interface water (triplicate—Water 1, Water 2, Water 3); (D) Control samples: an artificial sterile ice core made in the laboratory referred as a processing control, and a clean filter inside a clean microtube used as a control for downstream DNA analysis referred as DNA extraction control.